Tutorial 6: Hierarchical Path Integration Network¶
Tutorial Info
Reading Time: 35-40 minutes
Difficulty: Advanced
Prerequisites: Tutorial 1: Building and Using CANN Models
This tutorial introduces hierarchical path integration networks, which combine multi-scale grid cells [2] for robust spatial navigation in large environments.
Important
Reference Publication
This tutorial implements the model from: Chu et al. (2025)—“Localized Space Coding and Phase Coding Complement Each Other to Achieve Robust and Efficient Spatial Representation” [19]
For detailed theoretical background and experimental validation, please refer to the original paper.
1. Introduction to Hierarchical Path Integration¶
1.1 What is Hierarchical Path Integration?¶
Path integration is the ability to track position by integrating self-motion signals (velocity) over time. Hierarchical path integration [4, 19, 28] uses multiple grid cell [2] modules operating at different spatial scales to achieve:
Multi-scale representation: Coarse scales for large spaces, fine scales for precision
Error correction: Multiple scales provide redundancy against drift
Efficient coding: Different modules tile space at different resolutions
1.2 Biological Inspiration¶
In the mammalian brain:
Grid cells [2] in medial entorhinal cortex fire at regular spatial intervals forming hexagonal patterns
Multiple modules exist with different grid spacings (30cm to several meters)
Band cells integrate velocity along preferred directions
Place cells [1] in hippocampus receive convergent input from grid cells, forming localized spatial representations [19]
1.3 Key Components¶
The hierarchical network consists of:
2. Model Architecture¶
2.1 Component Overview¶
[ ]:
from canns.models.basic import HierarchicalNetwork
# Create hierarchical network with 5 modules
hierarchical_net = HierarchicalNetwork(
num_module=5, # Number of grid modules (different scales)
num_place=30, # Place cells per dimension (30x30 = 900 total)
spacing_min=2.0, # Smallest grid spacing
spacing_max=5.0, # Largest grid spacing
module_angle=0.0 # Base orientation angle
)
2.2 Key Parameters¶
Parameter |
Type |
Description |
|---|---|---|
|
int |
Number of grid modules with different scales |
|
int |
Place cells per spatial dimension |
|
float |
Minimum grid spacing (finest scale) |
|
float |
Maximum grid spacing (coarsest scale) |
|
float |
Base orientation for grid modules |
Parameter Guidelines:
num_module=5: Good balance between coverage and computationnum_place=30: Provides 900 place cells for 5x5m environmentSpacing range should match environment size (larger spaces need larger
spacing_max)
2.3 Internal Structure¶
Each module contains:
3 BandCell networks at 0°, 60°, 120° orientations
1 GridCell network combining band cell outputs
Connections project to shared place cell population
4. Visualization and Analysis¶
4.1 Computing Firing Fields¶
Firing fields show where each neuron is active in the environment:
[7]:
from canns.analyzer.metrics.spatial_metrics import compute_firing_field, gaussian_smooth_heatmaps
from canns.analyzer.visualization import PlotConfig, plot_firing_field_heatmap
# Prepare data
loc = np.array(task.data.position)
width = 5
height = 5
M = int(width * 10)
K = int(height * 10)
T = grid_r.shape[0]
# Reshape arrays for firing field computation
grid_r = grid_r.reshape(T, -1)
band_x_r = band_x_r.reshape(T, -1)
band_y_r = band_y_r.reshape(T, -1)
place_r = place_r.reshape(T, -1)
# Compute firing fields
print("Computing firing fields...")
heatmaps_grid = compute_firing_field(np.array(grid_r), loc, width, height, M, K)
heatmaps_band_x = compute_firing_field(np.array(band_x_r), loc, width, height, M, K)
heatmaps_band_y = compute_firing_field(np.array(band_y_r), loc, width, height, M, K)
heatmaps_place = compute_firing_field(np.array(place_r), loc, width, height, M, K)
# Apply Gaussian smoothing
heatmaps_grid = gaussian_smooth_heatmaps(heatmaps_grid)
heatmaps_band_x = gaussian_smooth_heatmaps(heatmaps_band_x)
heatmaps_band_y = gaussian_smooth_heatmaps(heatmaps_band_y)
heatmaps_place = gaussian_smooth_heatmaps(heatmaps_place)
print(f"Firing fields computed: {heatmaps_grid.shape}")
Computing firing fields...
Firing fields computed: (2000, 50, 50)
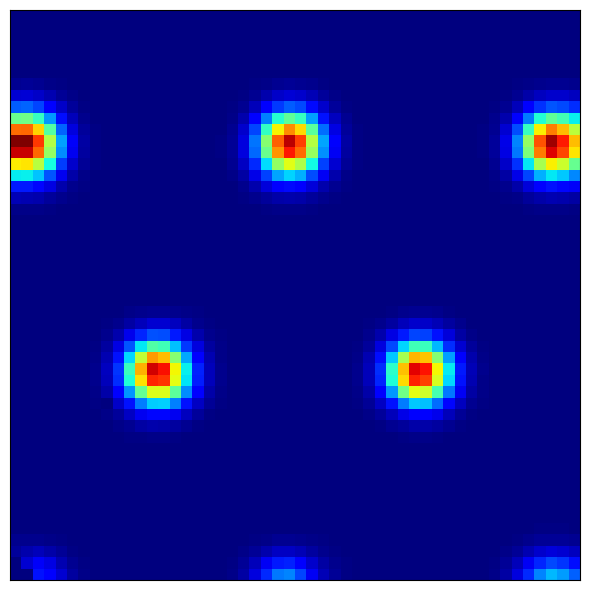
4.2 Visualizing Grid Cell Patterns¶
Grid cells show characteristic hexagonal firing patterns:
[9]:
# Reshape to separate modules
heatmaps_grid = heatmaps_grid.reshape(5, -1, M, K) # (modules, cells, x, y)
# Visualize a grid cell from module 0
module_idx = 0
cell_idx = 42
config = PlotConfig(
figsize=(6, 6),
title=f'Grid Cell - Module {module_idx}, Cell {cell_idx}',
xlabel='X Position (m)',
ylabel='Y Position (m)',
show=True,
save_path=None
)
plot_firing_field_heatmap(
heatmaps_grid[module_idx, cell_idx],
config=config
)
[9]:
(<Figure size 600x600 with 1 Axes>, <Axes: >)
What to observe:
Module 0 (finest scale): Small, tightly packed hexagons
Module 4 (coarsest scale): Large, widely spaced hexagons
Regular spacing: Grid vertices form equilateral triangles
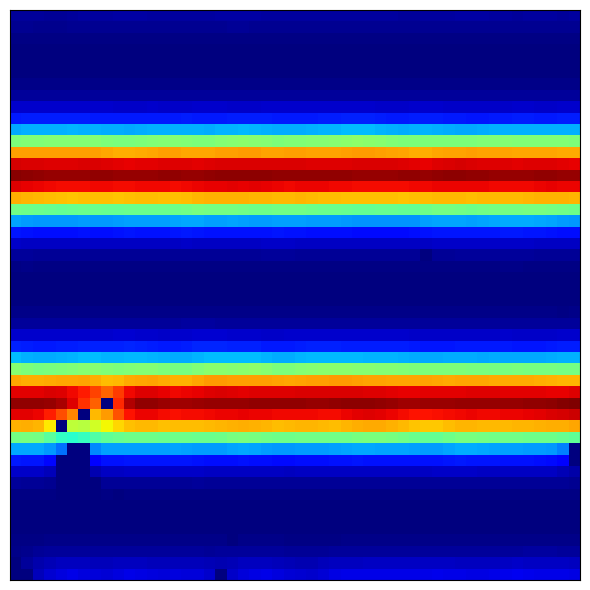
4.3 Visualizing Band Cell Patterns¶
Band cells show stripe-like patterns along their preferred orientation:
[10]:
# Reshape band cells
heatmaps_band_x = heatmaps_band_x.reshape(5, -1, M, K)
# Visualize a band cell
config = PlotConfig(
figsize=(6, 6),
title=f'Band Cell X - Module {module_idx}, Cell {cell_idx}',
xlabel='X Position (m)',
ylabel='Y Position (m)',
show=True,
save_path=None
)
plot_firing_field_heatmap(
heatmaps_band_x[module_idx, cell_idx],
config=config
)
[10]:
(<Figure size 600x600 with 1 Axes>, <Axes: >)
Expected patterns:
Parallel stripes perpendicular to preferred direction
Different cells have different stripe spacings (phase offsets)
Multiple modules show different scales of striping
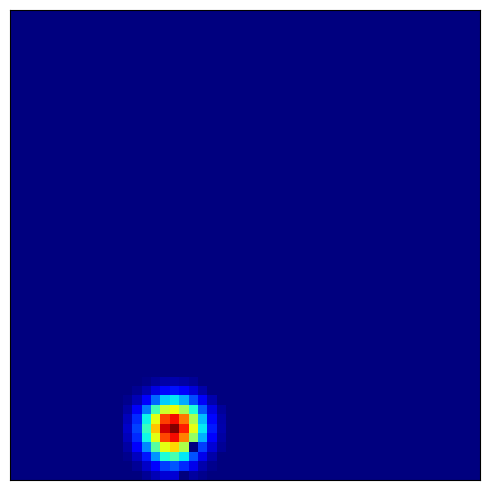
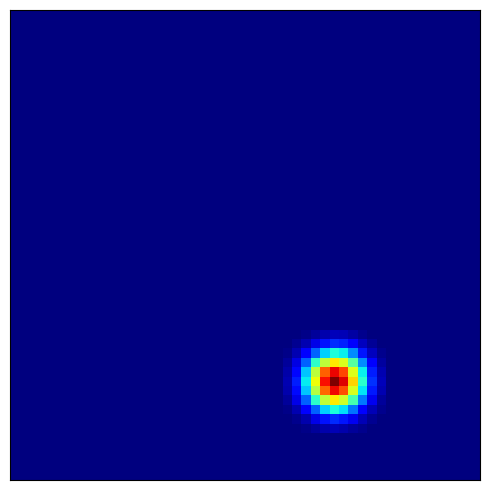
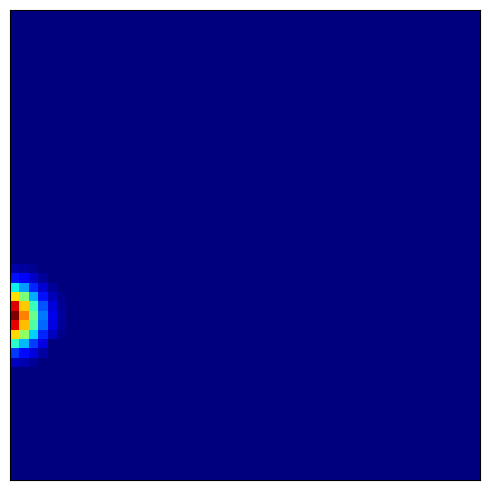
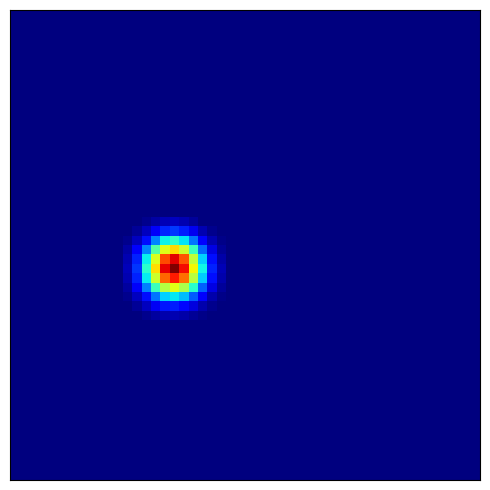
4.4 Visualizing Place Cell Fields¶
Place cells combine grid cell inputs to form localized firing fields:
[11]:
# Visualize several place cells
for place_idx in [100, 200, 300, 400]:
config = PlotConfig(
figsize=(5, 5),
title=f'Place Cell {place_idx}',
xlabel='X Position (m)',
ylabel='Y Position (m)',
show=True,
save_path=None
)
plot_firing_field_heatmap(
heatmaps_place[place_idx],
config=config
)
5. Next Steps¶
Congratulations! You’ve completed the hierarchical path integration tutorial. You now understand:
How multi-scale grid cell modules provide robust spatial coding
The architecture combining band cells, grid cells, and place cells
How to initialize hierarchical networks properly
How to visualize firing fields across different spatial scales
What You’ve Learned¶
- Hierarchical Architecture
You understand how multiple grid modules at different scales combine to create a robust spatial representation.
- Multi-Scale Coding
You’ve seen how coarse scales provide large-scale coverage while fine scales provide precision.
- Network Initialization
You know the critical importance of proper initialization with location input before running path integration.
- Biological Realism
You’ve implemented a model that matches the organization of the mammalian entorhinal-hippocampal system [2, 19].
Continue Learning¶
Explore related advanced topics:
Tutorial 7: Theta Sweep HD-Grid System - Learn theta-modulated systems combining head direction and grid cells
Tutorial 8: Theta Sweep Place Cell Network - Understand place cells in complex environments with geodesic distances
Or explore other scenarios:
Scenario 2: Data Analysis - Compare model predictions with experimental data
Scenario 3: Brain-Inspired Learning - Train spatial memory systems
Scenario 4: Pipeline - High-level pipeline tools for complete workflows
Key Takeaways¶
Multi-scale representation - Different grid spacings provide complementary information about position
Initialization matters - Strong location input is required to synchronize all modules
Band cells are building blocks - Grid cells emerge from combining band cell modules at different orientations
Place cells integrate - Place cells form localized fields by combining grid cell inputs
Advanced Topics¶
- Error Correction
Multiple scales allow the network to correct accumulated drift errors. Fine scales detect small errors, coarse scales provide global reference.
- Scaling Properties
Grid spacing typically follows a geometric progression (e.g., 2.0, 2.5, 3.1, 3.9, 4.9 meters for 5 modules).
- Biological Parameters
Real grid cells have spacings from ~30cm to several meters. Adjust
spacing_minandspacing_maxto match experimental data.- Place Field Formation
Place cells can develop multiple firing fields in large environments due to grid cell periodicity.
Parameter Tuning Guidelines¶
For hierarchical networks:
num_module=5-7: Good balance between coverage and computationspacing_min: Match finest experimental grid spacing (~30-50cm)spacing_max: Should cover environment size (e.g., 5m for 5x5m arena)num_place: Higher resolution for detailed place coding (30-50 per dimension)
Research Applications¶
This model is suitable for:
Path integration studies - How animals track position from self-motion
Grid cell research - Understanding multi-scale spatial coding
Hippocampal models - Linking entorhinal input to place cell formation
Navigation algorithms - Bio-inspired robotics and autonomous systems
5. Next Steps¶
Congratulations! You’ve completed the hierarchical path integration tutorial. You now understand:
How multi-scale grid cell modules provide robust spatial coding
The architecture combining band cells, grid cells, and place cells
How to initialize hierarchical networks properly
How to visualize firing fields across different spatial scales
What You’ve Learned¶
- Hierarchical Architecture
You understand how multiple grid modules at different scales combine to create a robust spatial representation.
- Multi-Scale Coding
You’ve seen how coarse scales provide large-scale coverage while fine scales provide precision.
- Network Initialization
You know the critical importance of proper initialization with location input before running path integration.
- Biological Realism
You’ve implemented a model that matches the organization of the mammalian entorhinal-hippocampal system [2, 19].
Continue Learning¶
Explore related advanced topics:
Tutorial 6: Theta Sweep HD-Grid System - Learn theta-modulated systems combining head direction and grid cells
Tutorial 7: Theta Sweep Place Cell Network - Understand place cells in complex environments with geodesic distances
Or explore other scenarios:
Scenario 2: Data Analysis - Compare model predictions with experimental data
Scenario 3: Brain-Inspired Learning - Train spatial memory systems
Scenario 4: Pipeline - High-level pipeline tools for complete workflows
Key Takeaways¶
Multi-scale representation - Different grid spacings provide complementary information about position
Initialization matters - Strong location input is required to synchronize all modules
Band cells are building blocks - Grid cells emerge from combining band cell modules at different orientations
Place cells integrate - Place cells form localized fields by combining grid cell inputs
Advanced Topics¶
- Error Correction
Multiple scales allow the network to correct accumulated drift errors. Fine scales detect small errors, coarse scales provide global reference.
- Scaling Properties
Grid spacing typically follows a geometric progression (e.g., 2.0, 2.5, 3.1, 3.9, 4.9 meters for 5 modules).
- Biological Parameters
Real grid cells have spacings from ~30cm to several meters. Adjust
spacing_minandspacing_maxto match experimental data.- Place Field Formation
Place cells can develop multiple firing fields in large environments due to grid cell periodicity.
Parameter Tuning Guidelines¶
For hierarchical networks:
num_module=5-7: Good balance between coverage and computationspacing_min: Match finest experimental grid spacing (~30-50cm)spacing_max: Should cover environment size (e.g., 5m for 5x5m arena)num_place: Higher resolution for detailed place coding (30-50 per dimension)
Research Applications¶
This model is suitable for:
Path integration studies - How animals track position from self-motion
Grid cell research - Understanding multi-scale spatial coding
Hippocampal models - Linking entorhinal input to place cell formation
Navigation algorithms - Bio-inspired robotics and autonomous systems
Next: Tutorial 6: Theta Sweep HD-Grid System