Design Philosophy & Architecture Overview¶
This document explains the core design principles and module organization of the CANNs library.
Core Design Principles¶
The CANNs library rests on two fundamental principles that guide its architecture and implementation.
Separation of Concerns¶
The library strictly separates different functional responsibilities into independent modules:
- 🏗️ Models (
canns.models): Define neural network dynamics and state evolution
- 📊 Tasks (
canns.task): Generate experimental paradigms and input data
- 📈 Analyzers (
canns.analyzer): Visualize and analyze simulation results
- 🧠 Trainers (
canns.trainer): Implement learning rules for brain-inspired models
- 🔗 Pipeline (
canns.pipeline): Orchestrate complete experimental workflows
Each module focuses on a single responsibility. Models don’t generate their own input data. Tasks don’t analyze results. Analyzers don’t modify model parameters. This separation makes the codebase maintainable, testable, and extensible.
Extensibility Through Base Classes¶
Every major component inherits from abstract base classes that define standard interfaces:
canns.models.basic.BasicModelfor basic CANN modelscanns.models.brain_inspired.BrainInspiredModelfor brain-inspired modelscanns.trainer.Trainerfor training algorithms
These base classes establish contracts ensuring all implementations work seamlessly with the rest of the library. Users can create custom models, tasks, or trainers by inheriting from these bases and implementing the required methods.
Module Architecture¶
Four Core Application Scenarios¶
The CANNs library supports four distinct workflows, each addressing different research needs. These scenarios demonstrate the modular design and flexibility of the architecture.
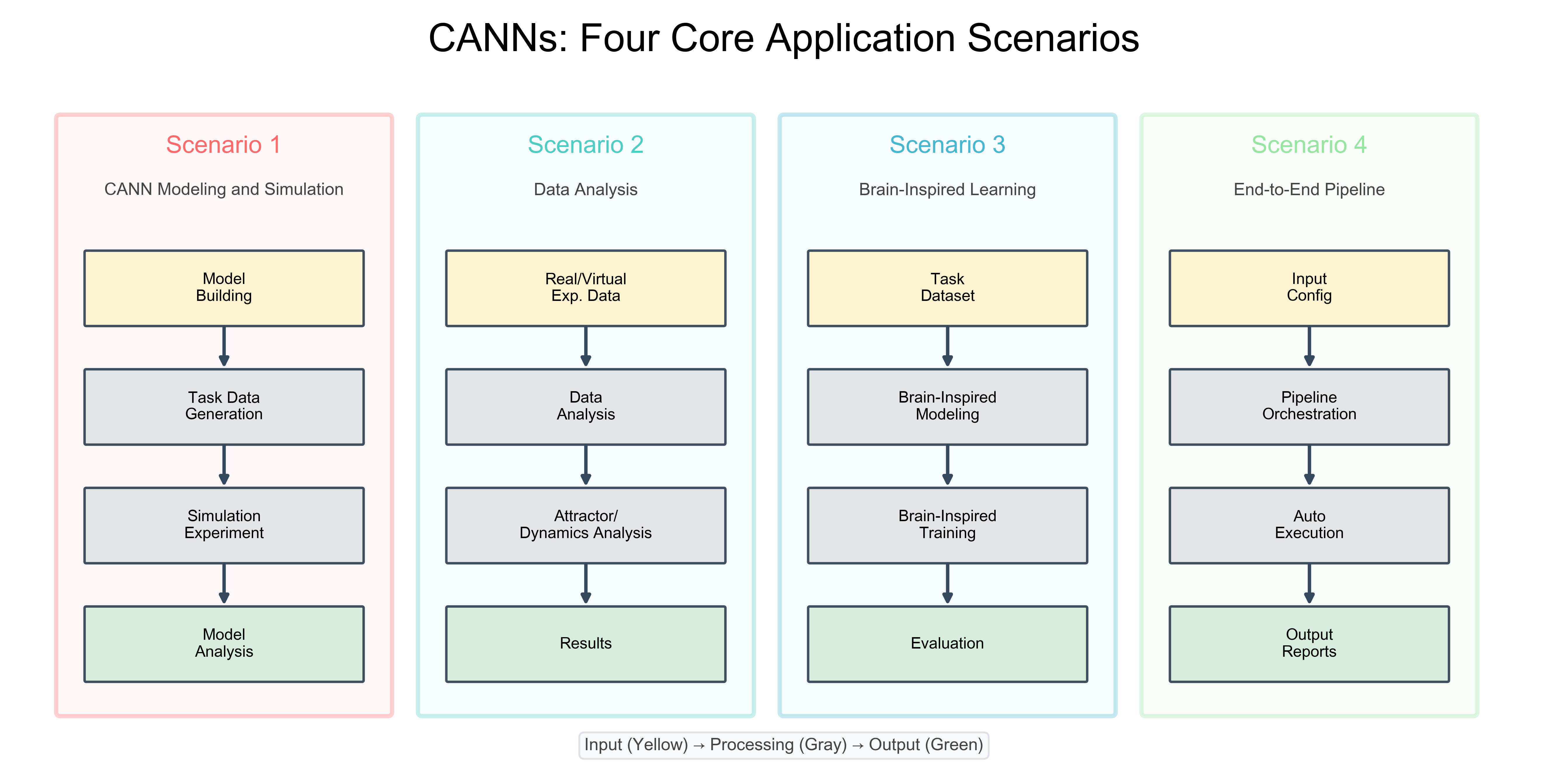
CANNs Four Core Application Scenarios¶
The most common workflow for studying continuous attractor dynamics
Model Building → Task Data Generation → Simulation Experiment → Model Analysis
Analyzing experimental or virtual neural recordings
Real/Virtual Exp. Data → Data Analysis → Attractor/Dynamics Analysis → Results
Training networks with biologically plausible learning rules
Task Dataset → Brain-Inspired Modeling → Brain-Inspired Training → Evaluation
Automated experimental workflows from configuration to results
Input Config → Pipeline Orchestration → Auto Execution → Output Reports
Module Interaction Pattern¶
Data Flow Pattern
Across all scenarios, modules interact following the separation of concerns principle:
🟡 Input Stage: Data enters the system (models, datasets, configs)
⚫ Processing Stage: Core computations (simulation, training, analysis)
🟢 Output Stage: Results visualization and interpretation
This consistent structure makes the library intuitive while supporting diverse research workflows.
BrainPy Integration¶
The CANNs library builds on BrainPy [18] (brainpy), a powerful framework for brain dynamics programming. BrainPy provides:
- ⚙️ Dynamics Abstraction:
bp.DynamicalSystembase class for neural systems- 💾 State Management:
bm.Variablecontainers for all state variables (replacing separate State, HiddenState, ParamState)- ⏱️ Time Step Control:
bm.set_dt(...)andbm.get_dt()for unified temporal management- ⚡ JIT Compilation:
bm.for_loopfor high-performance simulation- 🎲 Random Number Management:
bm.randomfor reproducible stochasticity
With BrainPy [18], CANN models only need to define variables and update equations. Time stepping, parallelization, and compilation are handled automatically—significantly reducing implementation complexity.
Module Relationships¶
How Modules Interact¶
Model ↔ Task Coupling¶
Some tasks require a model instance to access stimulus generation methods. For example, SmoothTracking1D needs access to model.get_stimulus_by_pos() to convert position coordinates into neural input patterns. This coupling is intentional for user convenience but is limited to tracking tasks.
Model ↔ Analyzer Independence¶
Analyzers work with model outputs (firing rates, membrane potentials) but don’t modify model state. They accept simulation results as NumPy arrays and produce visualizations. This independence allows the same analyzer to work with any model that produces compatible outputs.
Model ↔ Trainer Collaboration¶
Trainers modify model parameters—specifically connection weights—according to learning rules. They interact with models through agreed-upon attributes like model.W for weights and model.s for state vectors. The trainer framework is designed for brain-inspired models that use local, activity-dependent plasticity.
Pipeline Orchestration¶
The canns.pipeline module coordinates all other modules into complete experimental workflows. It manages the full cycle from model setup through task execution to result analysis, providing a high-level interface for common use cases.
Design Trade-offs¶
Flexibility vs. Convenience¶
Advanced users can override any component or create custom implementations
Standard workflows should require minimal boilerplate
The library achieves this balance through sensible defaults combined with extensive customization options. For example, CANN1D() uses default parameters that work for most cases, but every parameter can be explicitly specified.
Performance vs. Simplicity¶
The library achieves high performance through a multi-layered strategy:
Python Layer (BrainPy/JAX [17])¶
JAX [17]-based compilation provides GPU/TPU acceleration but requires functional programming patterns. The library abstracts this complexity by:
Encapsulating JIT compilation in BrainPy’s
bm.for_loopManaging state through
bm.VariablecontainersProviding utility functions that handle common patterns
Users benefit from GPU/TPU acceleration without writing JAX-specific code directly.
Native Layer (canns-lib)¶
Important
For performance-critical operations where Python overhead is significant, the library provides optional Rust-powered backends through canns-lib:
Ripser Module: Topological Data Analysis [15, 16] with 1.13x average speedup (up to 1.82x) vs. pure Python
Spatial Navigation: Accelerated RatInABox environments with ~700x speedup for long trajectory integration
Future Modules: Planned support for approximate nearest neighbors, dynamics computation
The canns-lib integration follows the same principle: it exposes simple Python APIs while leveraging native performance for bottleneck operations. Users can opt into these accelerations without changing their code structure.
Extending the Library¶
Creating Custom Models¶
To add a new model, inherit from the appropriate base class and implement required methods.
Required methods:
make_conn(): Generate connection matrixget_stimulus_by_pos(): Convert positions to input patternsinit_state(): Register state variablesupdate(): Define single-step dynamics
Required methods:
init_state(): Register state and weight parametersupdate(): Define state evolutionenergy: Property returning network energy
Creating Custom Tasks¶
Tip
Tasks should generate input sequences compatible with model expectations. Key considerations:
Use
bm.get_dt()for time step consistencyReturn data in formats expected by models
Provide trajectory information for analysis
Creating Custom Trainers¶
Note
Trainers inherit from canns.trainer.Trainer and implement:
train(): Parameter update strategypredict(): Single-sample inferenceStandard progress and compilation configuration
Summary¶
The CANNs library achieves its goals through careful architectural choices:
1️⃣
Separation of concerns keeps modules focused and independent
2️⃣
Base class inheritance ensures consistent interfaces
3️⃣
BrainPy integration provides performance without complexity
4️⃣
Flexible coupling balances convenience with modularity
These principles enable both rapid prototyping and rigorous research while maintaining code quality and extensibility.